The Mouse Biology Program (MBP) of the University of California, Davis (UCD) is the largest academic program in the world. MBP is a member of the NIH-funded Mutant Mouse Resource and Research Centers (MMRRC). The network collects and characterizes gene-modified and genetic knockout lab mice and makes them available to 800 researchers at over 400 institutions in 42 countries around the globe. [1]
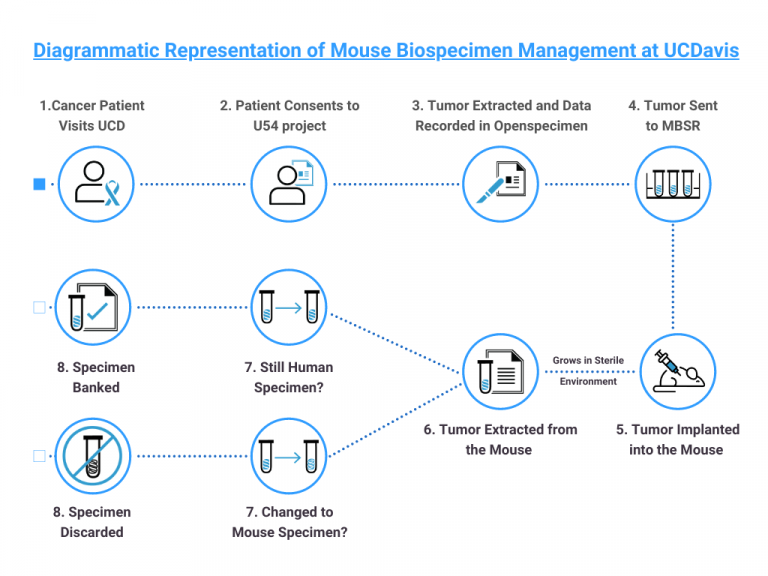
Tumor samples are surgically resected from participants consenting to the U54 study and implanted to mouse models in MBSR (Mouse Biology Shared Resources). The biospecimen is allowed to grow in the mouse in a sterile controlled environment and are sent back to UCD for QA (Immunohistochemistry and KI67 staining).
The data is recorded in OpenSpecimen throughout the process.
Implementation in OpenSpecimen
OpenSpecimen has a highly configurable user interface that can be designed as per the study’s needs. It is possible to configure studies to record both human and animal biospecimen data in the same instance.
OpenSpecimen Screenshots
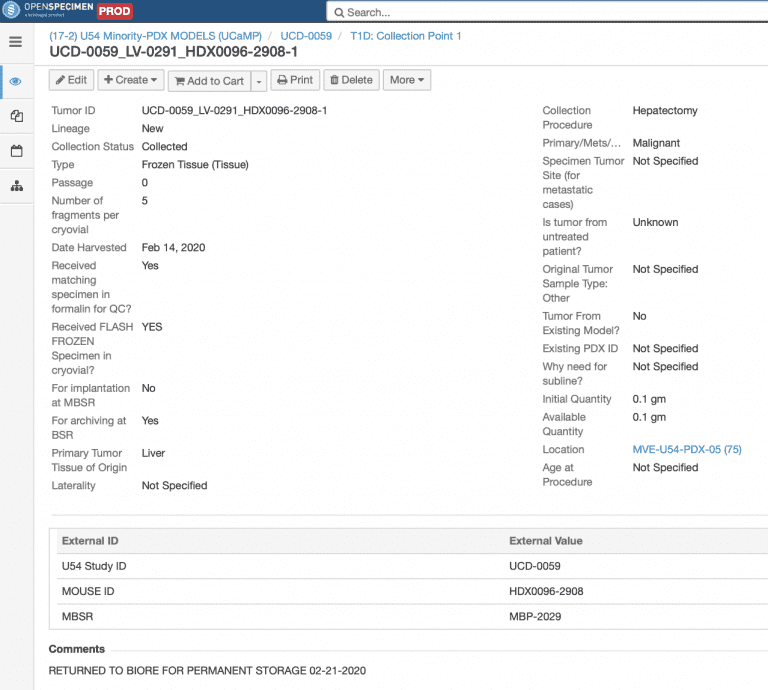
Tumor Details:
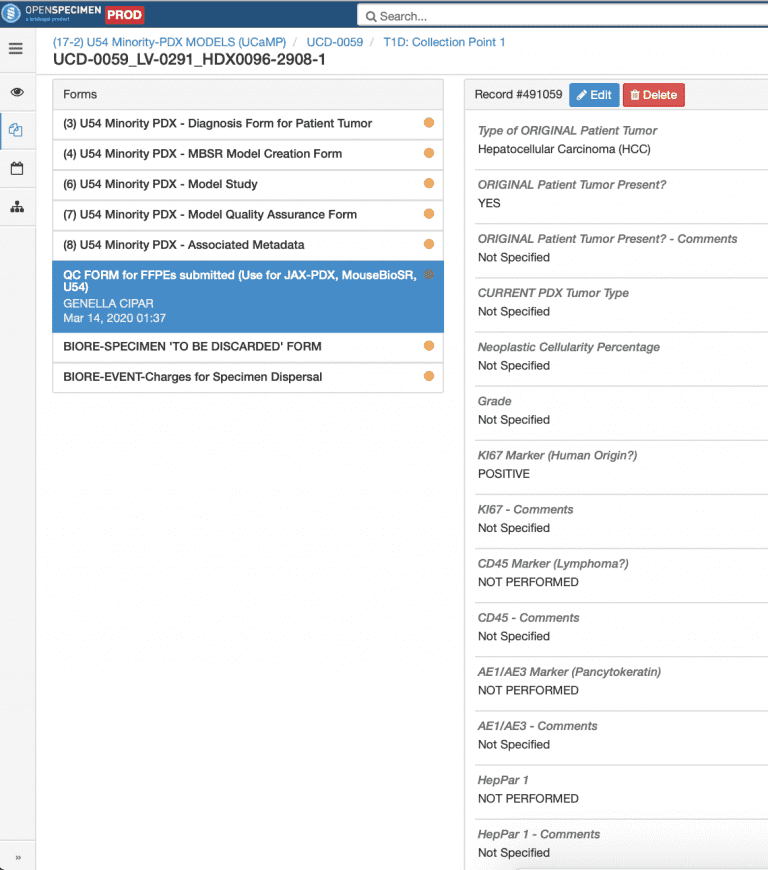
QA Data:
Reporting:
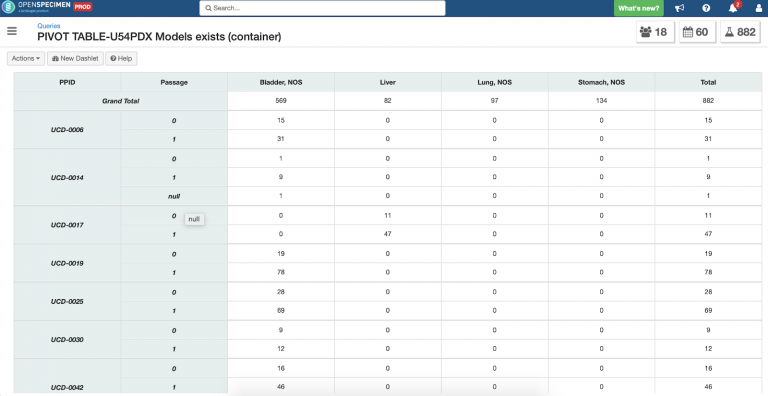
Using OpenSpecimen’s reporting interface, it’s straightforward to retrieve data for these specimens/mouse models or the U54 participants.
Example pivot table report to g
Check the staining results and markers:
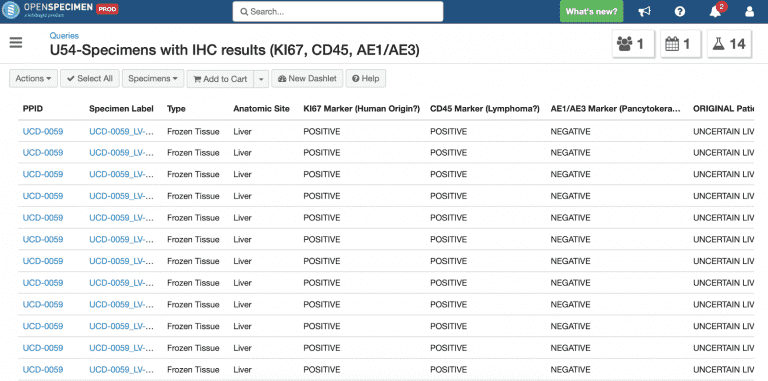
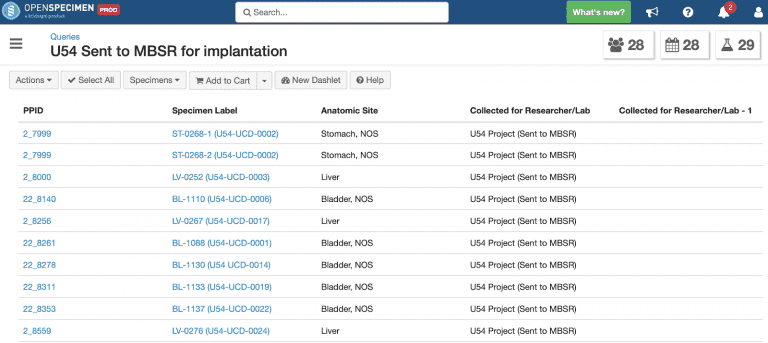
Check specimens extracted from patients and sent to MBSR for mouse implantation:
Summary
“We could not have tracked the outgoing U54 participant specimens and the incoming U54 PDX models without OpenSpecimen’s high degree of configurability. Data entry is smooth and straightforward. For easy access to data, I have set up pivot tables showing the number of specimens per PDX model plus tumor site and passage.” said Irmgard Feldman, Biorepository Manager at the Department of Pathology and Laboratory Medicine at UC Davis.
References:
https://www.youtube.com/watch?v=935_Svv0yXg
https://www.mousebiology.org/component/content/featured
https://health.ucdavis.edu/cancer/research/sharedresources/mouse.html
Written by: Neha Nimgire, Senior Member of Domain Staff, Krishagni
For more details, email [email protected]
